Publication Detail
Reassortment between divergent strains of Camp Ripley virus (Hantaviridae) in the northern short-tailed shrew (Blarina brevicauda)
Liphardt SW, Kang HJ, Arai S, Gu SH, Cook JA, Yanagihara R
Citation
Liphardt SW, Kang HJ, Arai S, Gu SH, Cook JA, Yanagihara R (2020) Reassortment between divergent strains of Camp Ripley virus (Hantaviridae) in the northern short-tailed shrew (Blarina brevicauda) Frontiers in Cellular and Infection Microbiology 10, 460
Abstract
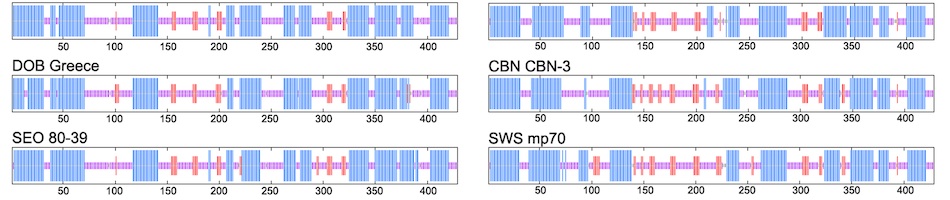
Genomic reassortment of segmented RNA virus strains is an important evolutionary mechanism that can generate novel viruses with profound effects on human and animal health, such as the H1N1 influenza pandemic in 2009 arising from reassortment of two swine influenza viruses. Reassortment is not restricted to influenza virus and has been shown to occur in members of the order Bunyavirales. The majority of reassortment events occurs between closely related lineages purportedly due to molecular constraints during viral packaging. In the original report of Camp Ripley virus (RPLV), a newfound hantavirus in the northern short-tailed shrew (Blarina brevicauda), phylogenetic incongruence between different genomic segments suggested reassortment. We have expanded sampling to include RPLV sequences amplified from archival tissues of 36 northern short-tailed shrews collected in 12 states (Arkansas, Iowa, Kansas, Maryland, Massachusetts, Michigan, Minnesota, New Hampshire, Ohio, Pennsylvania, Virginia, Wisconsin), and one southern short-tailed shrew (Blarina carolinensis) from Florida, within the United States. Using Bayesian phylogenetic analysis and Graph-incompatibility-based Reassortment Finder, we identified multiple instances of reassortment that spanned the Hantaviridae phylogenetic tree, including three highly divergent, co-circulating lineages of the M segment that have reassorted with a conserved L segment in multiple populations of B. brevicauda. In addition to identifying the first known mobatvirus-like M-segment sequences from a soricid host and only the second from a eulipotyphlan mammal, our results suggest that reassortment may be common between divergent virus strains and provide strong justification for expanded spatial, temporal, and taxonomic analyses of segmented viruses.
| Link: | https://pubmed.ncbi.nlm.nih.gov/33014888/ |
| PMID: | |
| PMCID: | PMC7509084 |