Publication Detail
Virus DNA-fragment classification using taxonomic hidden Markov model profile.
Menor M, Baek K, Belcaid M, Gingras Y, Poisson G.
Citation
Menor M, Baek K, Belcaid M, Gingras Y, Poisson G. (2010) Virus DNA-fragment classification using taxonomic hidden Markov model profile. Proceedings of the 2010 ACM Symposium on Applied Computing 1567-1571.
Abstract
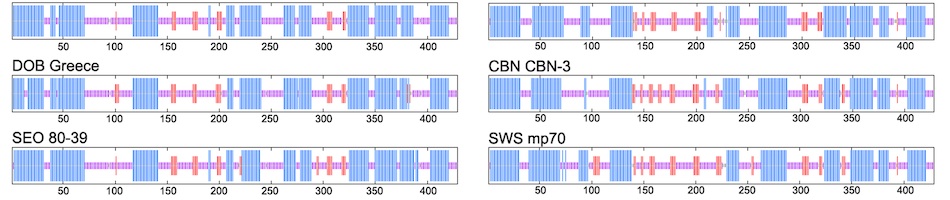
In most viral metagenomic studies, genetic material from a diversity of organisms is sampled from the environment and sequenced using Sanger or 454 sequencing. This process typically results in DNA-fragments that need to be assembled into contigs and annotated before any inferences or conclusions can be drawn from the data in hand. However, one problem subsists: both the relatively short length of the sequenced DNA-fragments and the high level of diversity present in a viral community result in a large number of unassembled and unannotated DNA-fragments. This problem limits our capability to better understand the viral community under study. We present the preliminary results of a new annotation method, targeting the virus sequences highly likely to be left unannotated by conventional methods. The resulting system, called Anacle, gives a taxonomic annotation for virus sequences excluded by a pre-screening with BLAST. Anacle uses an automated method relying a) on the Markov clustering (MCL) of all protein sequences belonging to the same taxon and b) on constructing each taxon’s genetic variations (skeletons) using Hidden Markov Model (HMM) profiles. The taxonomic annotation consists of comparing each unannotated DNA-fragment to all the skeletons, and labeling them as belonging to the taxon associated with the best similarity score. We have evaluated Anacle’s performance on a simulated metagenomes dataset with 100 and 700 base pairs. The results show that Anacle can taxonomically annotate viruses DNA-fragments with high precision and specificity. It indicates that the proposed method can provide valuable taxonomic information about DNA-fragments that could be left unannotated by other methods. We also present Anacle’s performance on a small Sargasso Sea dataset.
| Link: | http://portal.acm.org/citation.cfm?id=1774423 |
| PMID: | |
| PMCID: |