Publication Detail
Biospecimen repositories and integrated databases as critical infrastructure for pathogen discovery and pathobiology research.
Dunnum JL, Yanagihara R, Johnson KM, Armien B, Batsaikhan N, Morgan L, Cook JA.
Citation
Dunnum JL, Yanagihara R, Johnson KM, Armien B, Batsaikhan N, Morgan L, Cook JA. (2017) Biospecimen repositories and integrated databases as critical infrastructure for pathogen discovery and pathobiology research. PLoS Neglected Tropical Diseases 11:e0005133.
Abstract
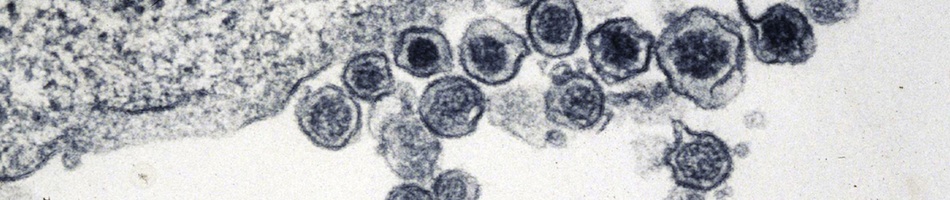
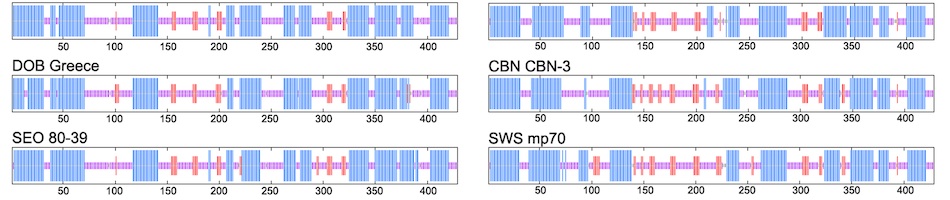
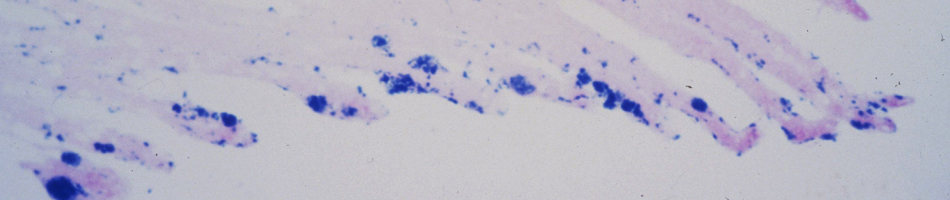
Field collections of natural history specimens often arise through dynamic collaborations that are capable of producing a diverse array of preparations and associated data (e.g., ultra-frozen tissue, cell suspensions, feces, and endo- and ecto-parasites) with precise spatial and temporal stamps that facilitate myriad investigations. When properly archived and digitally captured, museum databases are capable of linking diverse kinds of “big data”. This biorepository nexus can be a powerful tool for research in pathogen discovery, environmental change, and host-reservoir dynamics. Spatially broad and temporally deep archives of ultra-frozen tissues represent unparalleled infrastructure for virologists, as demonstrated through the retrospective surveys for Sin Nombre hantavirus and subsequent significant new hantavirus discoveries across four continents. As tools for extracting vast amounts of information from both contemporary and ancient specimens improve, new insights into pathogen evolution and ecology will be enhanced. We suggest that the benefits of incorporating this model into pathogen discovery and pathobiology research far outweigh any potential costs associated with its implementation.
| Link: | https://www.ncbi.nlm.nih.gov/pubmed/28125619 |
| PMID: | 28125619 |
| PMCID: | PMC5268418 |