Publication Detail
Genetic analysis of Hokkaido hantavirus among Myodes rufocanus in the Baikal Lake area.
Yashina LN, Danchinova GA, Seregin SV, Khasnatinov MA, Yanagihara R.
Citation
Yashina LN, Danchinova GA, Seregin SV, Khasnatinov MA, Yanagihara R. (2013) Genetic analysis of Hokkaido hantavirus among Myodes rufocanus in the Baikal Lake area. Bulletin of Eastern Siberian Scientific Center 4(92):147-152.
Abstract
Background: Hokkaido hantavirus (HOKV) was identified originally in the grey red-backed vole (Myodes rufocanus) in Hokkaido, Japan. Subsequent studies showed different genetic lineages of HOKV in Sakhalin, Buryatia and Far Eastern regions of Russia and in China.
Methods: Tissues from 68 arvicolid rodents, captured in regions south and west of Baikal Lake, were initially tested for hantaviral antigens by ELISA, and tissues from antigen-positive rodents were analyzed for hantavirus RNA by RT-PCR. Taxonomic identification of host species was based on phylogenetic analysis of partial cytochrome b gene sequences.
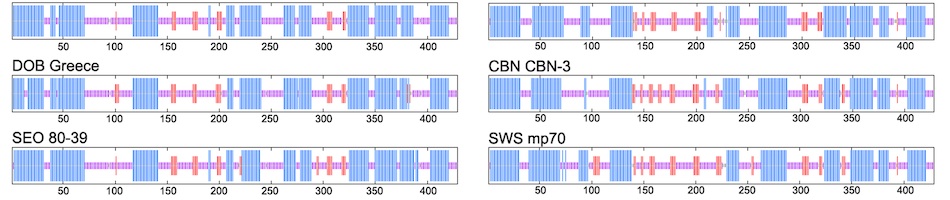
Results: Hantavirus L- and S-segment sequences were detected in two antigen-positive M. rufocanus, from the Tunka region of Buryatia Republic (south side) and the Olhon region of Irkutsk Oblast (west side). Sequence analysis showed that the newfound hantavirus strains, designated Baikal and Siberia, represented genetic variants of HOKV. Previously unknown genetic variant designated Siberia was identified in M. rufocanus captured in Olhon region. A second genetic variant from Tunka region, designated Baikal, was closely related to previously described hantavirus strain from the same region. Alignment and comparison of the nucleotide and amino acid sequences showed intra-strain differences of 18.4% and 5.3% for the L segment and 17.4% and 3.5% for the S segment, respectively. Sequence divergence from geographically distant HOKV strains were 17.4�¢??21.5% and 3.9�¢??6.8% for the L segment and 15.2�¢??17.0% and 3.3�¢??4.0% for the S segment, respectively. Phylogenetic analysis, based on a 346-nucleotide region of the L segment, revealed
four lineages represented by previously reported variants from Japan and Sakhalin (strains Kitahiyama128L/2008, Tobetsu35L/2010 and Sakhalin99L/1998), Shkotovo in Far-Eastern Russia (strain Khekhtsir37L/2002) and the new variants from Baikal Lake. Analysis of the N protein, coding by the S segment, identified specific amino acid signatures for HOKV of Lys5, Arg26, Val/Ile68, Val/Ala79, Ile262, Pro283.
Conclusions: HOKV is widespread across the geographic range of its arvicolid rodent reservoir host.
| Link: | |
| PMID: | |
| PMCID: |