Publication Detail
Hantaviruses: rediscovery and new beginnings.
Yanagihara R, Gu SH, Arai S, Kang HJ, Song JW.
Citation
Yanagihara R, Gu SH, Arai S, Kang HJ, Song JW. (2014) Hantaviruses: rediscovery and new beginnings. Virus Research 187:6-14.
Abstract
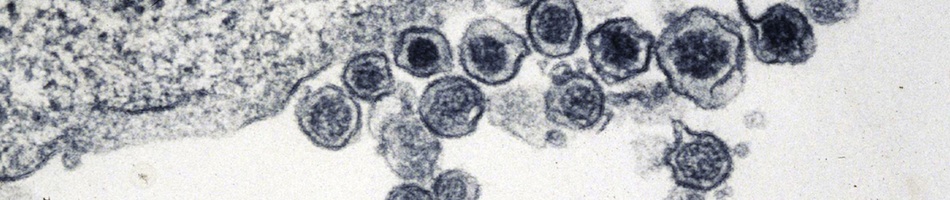
Virus and host gene phylogenies, indicating that antigenically distinct hantaviruses (family Bunyaviridae, genus Hantavirus) segregate into clades, which parallel the molecular evolution of rodents belonging to the Murinae, Arvicolinae, Neotominae and Sigmodontinae subfamilies, suggested co-divergence of hantaviruses and their rodent reservoirs. Lately, this concept has been vigorously contested in favor of preferential host switching and local host-specific adaptation. To gain insights into the host range, spatial and temporal distribution, genetic diversity and evolutionary origins of hantaviruses, we employed reverse transcription- polymerase chain reaction to analyze frozen, RNAlater-preserved and ethanol-fixed tissues from 1,546 shrews (9 genera, 47 species), 281 moles (8 genera, 10 species) and 520 bats (26 genera and 53 species), collected in Europe, Asia, Africa and North America during 1980-2012. Thus far, we have identified 24 novel hantaviruses in shrews, moles and bats. That these newfound hantaviruses are geographically widespread and genetically more diverse than those harbored by rodents suggests that the evolutionary history of hantaviruses is far more complex than previously conjectured. Phylogenetic analyses indicate four distinct clades, with the most divergent comprising hantaviruses harbored by the European mole and insectivorous bats, with evidence for both co-divergence and host switching. Future studies will provide new knowledge about the transmission dynamics and pathogenic potential of these newly discovered, still-orphan, non-rodent-borne hantaviruses.
| Link: | http://www.ncbi.nlm.nih.gov/pubmed/24412714 |
| PMID: | 24412714 |
| PMCID: | PMC4058404 |