Publication Detail
Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides).
Arai S, Ohdachi SD, Asakawa M, Kang HJ, Mocz G, Arikawa J, Okabe N, Yanagihara R.
Citation
Arai S, Ohdachi SD, Asakawa M, Kang HJ, Mocz G, Arikawa J, Okabe N, Yanagihara R. (2008) Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides). Proceedings of the National Academy of Sciences USA 105(42):16296-16301.
Abstract
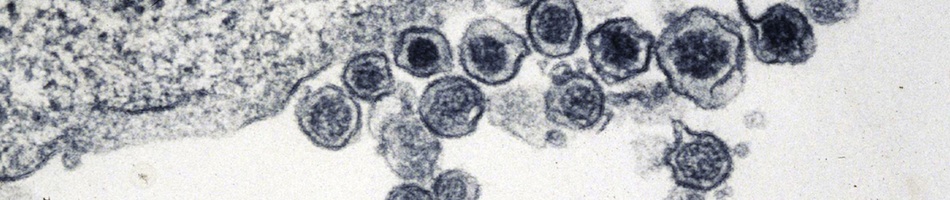
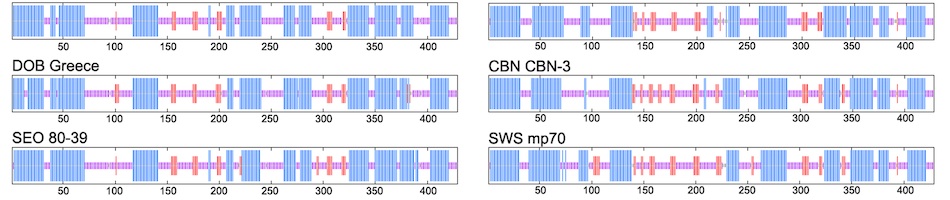
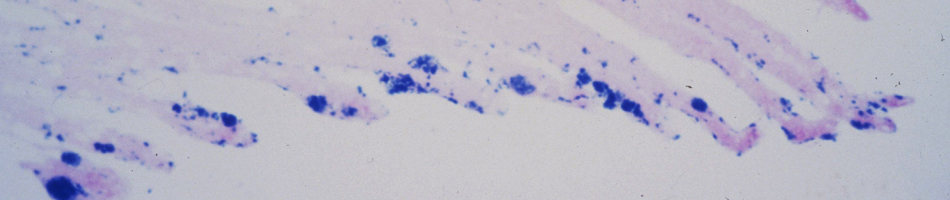
Recent molecular evidence of genetically distinct hantaviruses in shrews, captured in widely separated geographical regions, corroborates decades-old reports of hantavirus antigens in shrew tissues. Apart from challenging the conventional view that rodents are the principal reservoir hosts, the recently identified soricid-borne hantaviruses raise the possibility that other soricomorphs, notably talpids, similarly harbor hantaviruses. In analyzing RNA extracts from lung tissues of the Japanese shrew mole (Urotrichus talpoides), captured in Japan between February and April 2008, a hantavirus genome, designated Asama virus (ASAV), was detected by RT-PCR. Pairwise alignment and comparison of the S-, M-, and L-segment nucleotide and amino acid sequences indicated that ASAV was genetically more similar to hantaviruses harbored by shrews than by rodents. However, the predicted secondary structure of the ASAV nucleocapsid protein was similar to that of rodent- and shrew-borne hantaviruses, exhibiting the same coiled-coil helix at the amino terminus. Phylogenetic analyses, using the maximum-likelihood method and other algorithms, consistently placed ASAV with recently identified soricine shrew-borne hantaviruses, suggesting a possible host-switching event in the distant past. The discovery of a mole-borne hantavirus enlarges our concepts about the complex evolutionary history of hantaviruses.
| Link: | http://www.ncbi.nlm.nih.gov/pubmed/18854415 |
| PMID: | 18854415 |
| PMCID: | PMC2567236 |